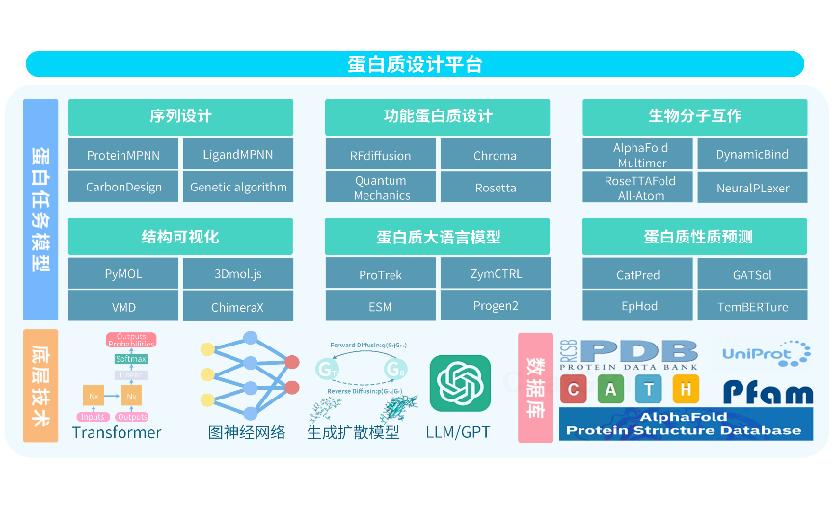
Protein Design Pipeline
A comprehensive protein design pipeline integrating AI, structure visualization, and molecular interaction tools.
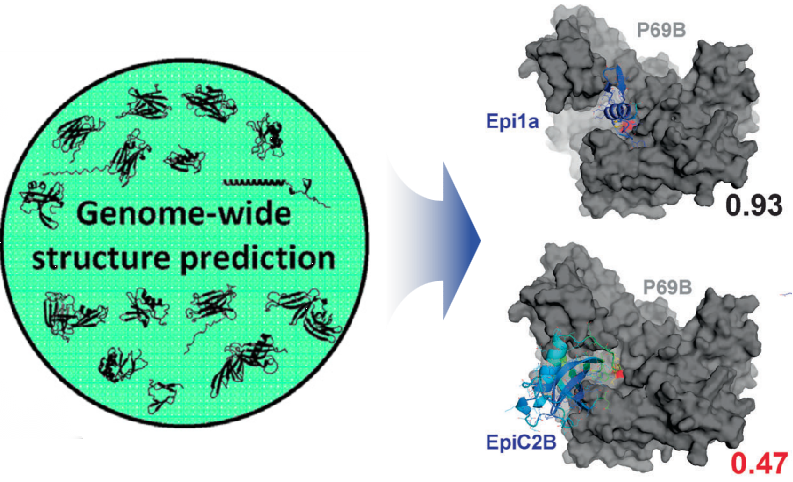
Effector Prediction Pipeline
Predict effector proteins and target from microbial genomes.

Virtual Screening Pipeline
A multi-step virtual screening pipeline from database searches of billions of compounds to detailed molecular simulations.
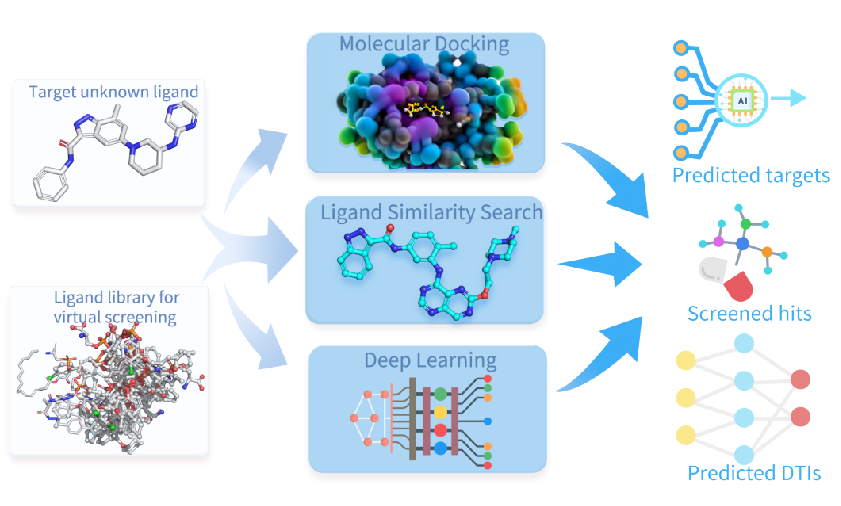
Target Fishing Pipeline
An AI-powered target fishing pipeline combining molecular docking, ligand similarity search, and deep learning to predict targets, screen hits, and identify drug-target interactions.
AlphaFold2
Accurately predict protein and complex structures at the atomic level using their amino acid sequence.
Chai-1
Chai-1 replicates AlphaFold3's capabilities, enabling accurate prediction of biomolecular structures.

HelixFold3
HelixFold3 replicates AlphaFold3's capabilities, enabling accurate prediction of biomolecular structures.

FastGPT
A platform for streamlined LLM-powered knowledge management and deployment.
ESMFold
Accurately predict protein structures at the atomic level using its amino acid sequence.

RoseTTAFold2
Protein structure prediction that's faster than AlphaFold2 and just as accurate.
RFdiffusion-v2
Design proteins, binders, and more with this protein diffusion model.
RoseTTAFold All-Atom
Protein folding model that supports proteins, nucleotides, ligands, metal ions, and other small molecules.
LigandMPNN
Predict alternative sequences for an input protein structure with high accuracy. Also supports ProteinMPNN and SolubleMPNN.

ProteinMPNN
Predict alternative sequences for an input protein structure with high accuracy. Also supports SolubleMPNN.

ProGen2
Create protein variants using nothing but the amino acid sequence.
AF2Bind
Accurately predict small-molecule-binding residues using AlphaFold2 pairwise representation.
DynamicBind
Predict protein-ligand complexes using protein structure files and ligands in SMILES format.
DiffDock-L
Dock a ligand onto any protein receptor with high accuracy.
PocketFlow
PocketFlow is a Deep Generative Model that generates ligands for target protein binding pockets.
AlphaFlow
Use AlphaFlow to generate protein structures that closely reflect experimental and physiological conditions.
DLKcat Kcat Prediction
Predict Kcats of complexes using a protein sequence and compounds in SMILES format.
EpHod Optimal Enzyme pH Prediction
EpHod is a semi-supervised language model that predicts optimal pH for enzymes from sequence alone.
Conformer Generator
Generate conformers for small molecules and ligands using RDKit.
LightDock
Powerful molecular docking algorithm for proteins and nucleotides.

ESM-2 for PTMs
Predict potential post-translational modification sites.
























